easyunfold Documentation#
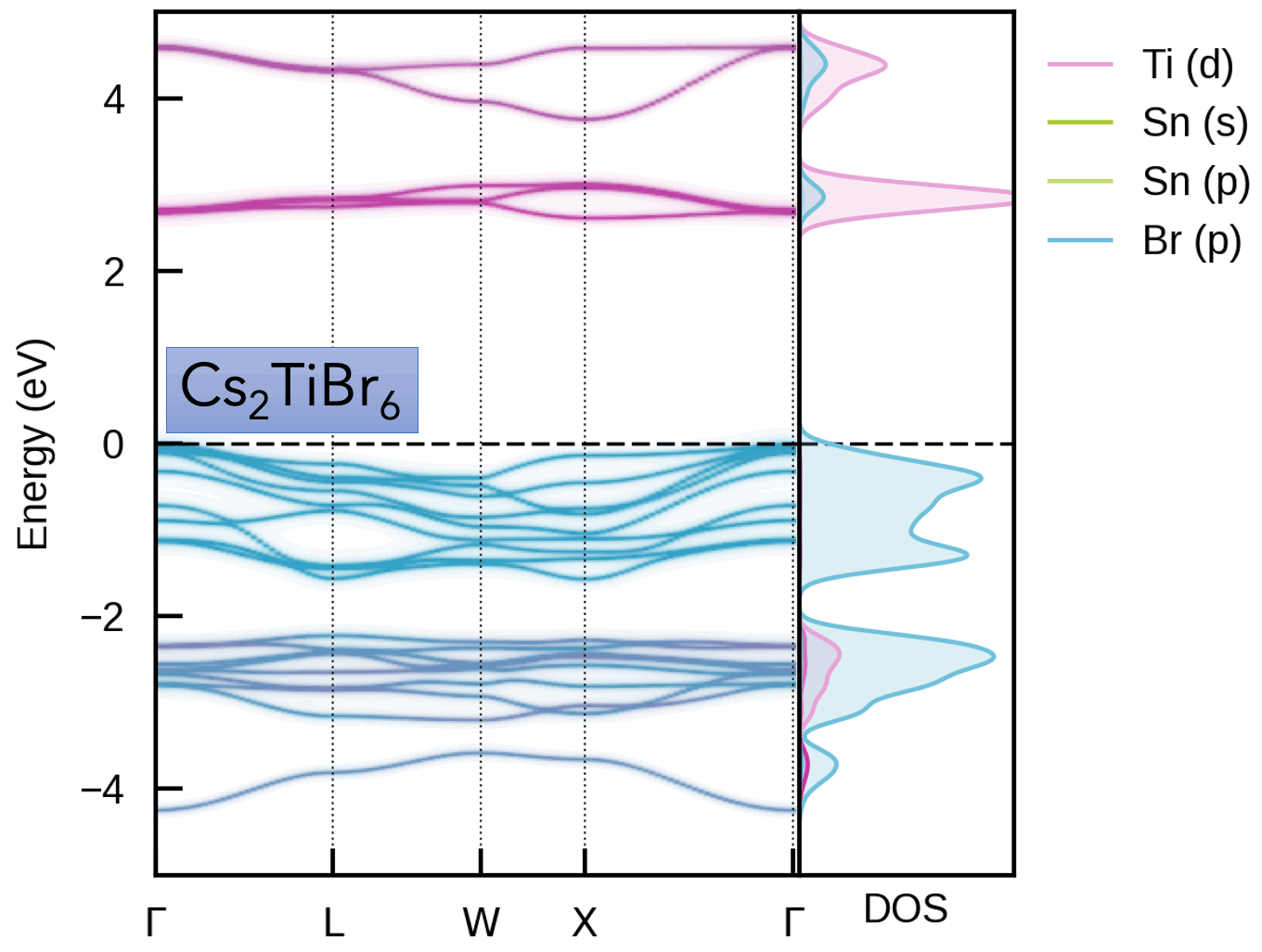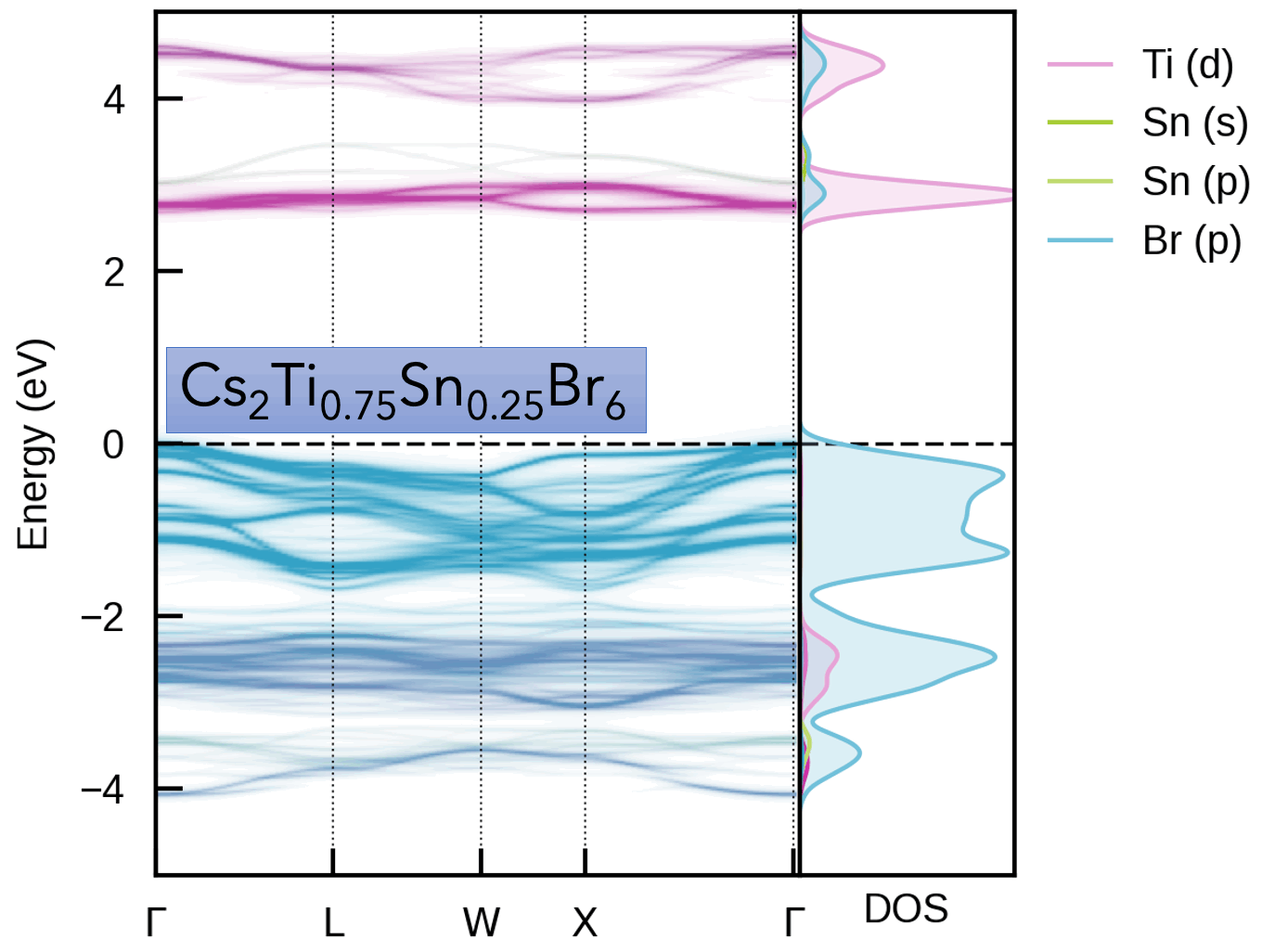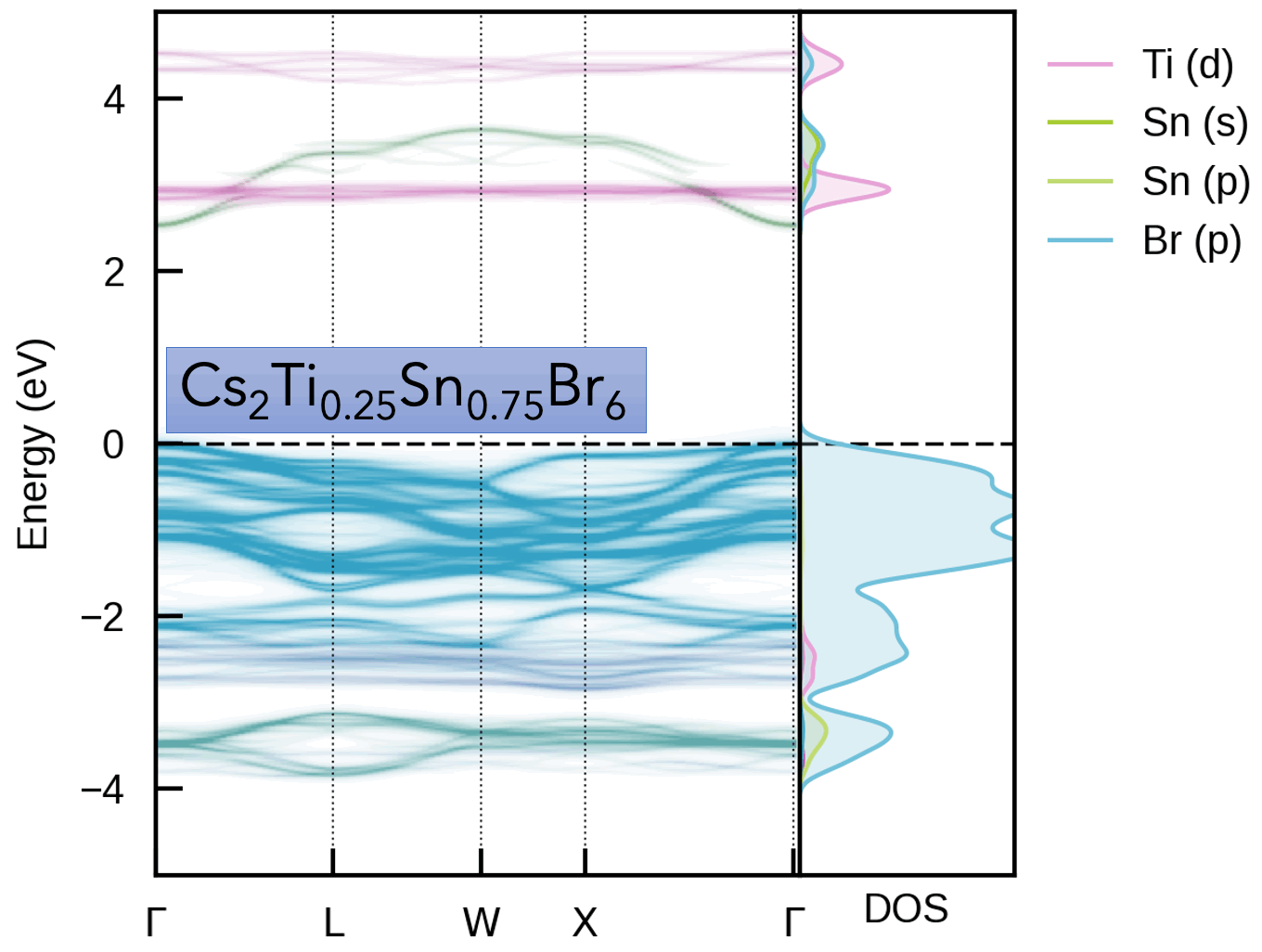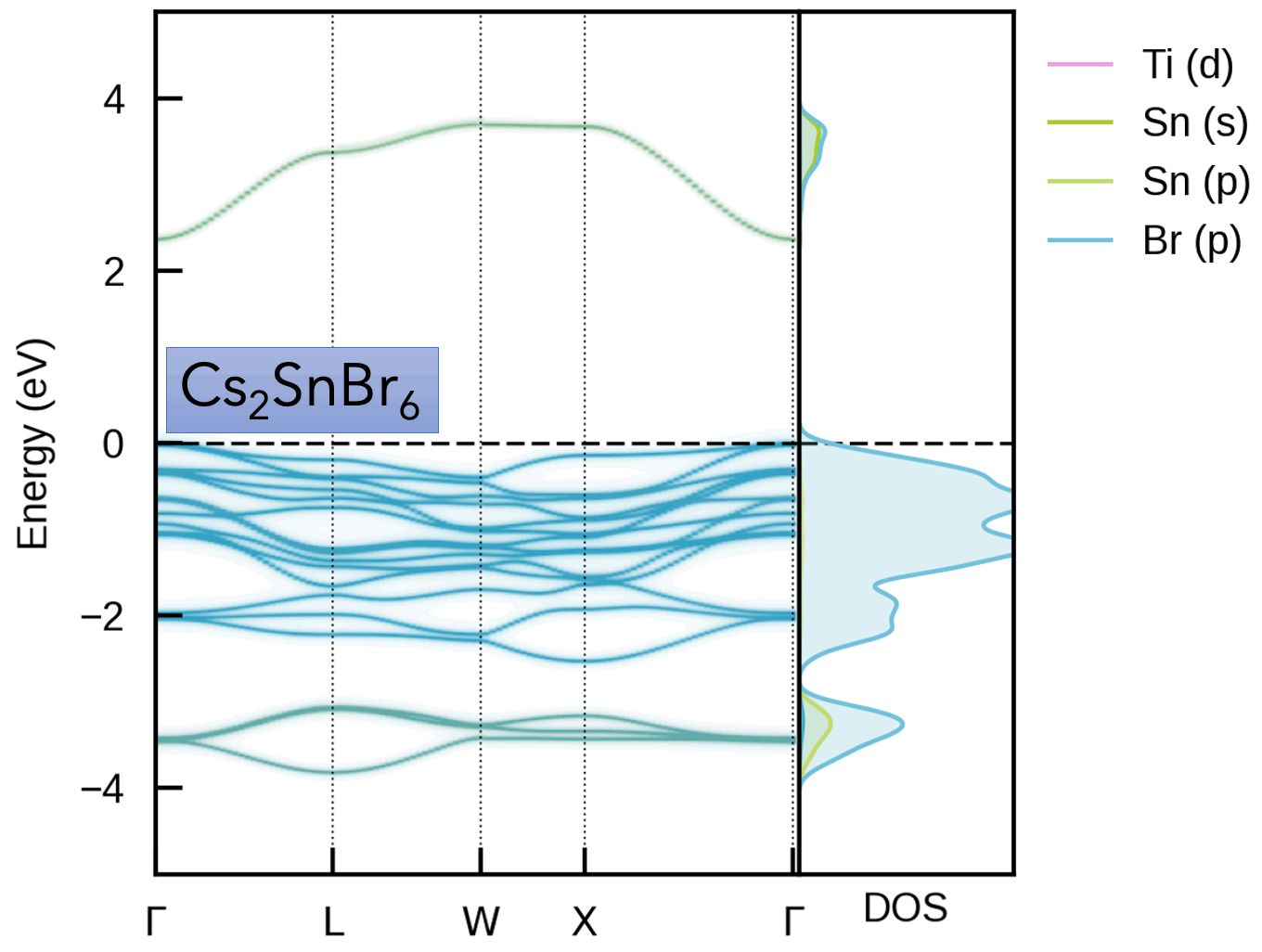
This package is intended for obtaining the effective band structure of a supercell for a certain k-point path of the primitive cell. It was originally based on PyVaspwfc for reading VASP wavefunction outputs, with a notable improvement being that symmetry-breaking is properly accounted for by sampling necessary additional k-points and averaging accordingly. Typical applications of band structure unfolding are the electronic structure analysis of defects, disorder, alloys, surfaces (and more), as illustrated in the example outputs below and docs examples.
Our goal is to implement the band structure unfolding workflow in a robust and user-friendly software package.
For the methodology of supercell band unfolding, see here.
Example Outputs#
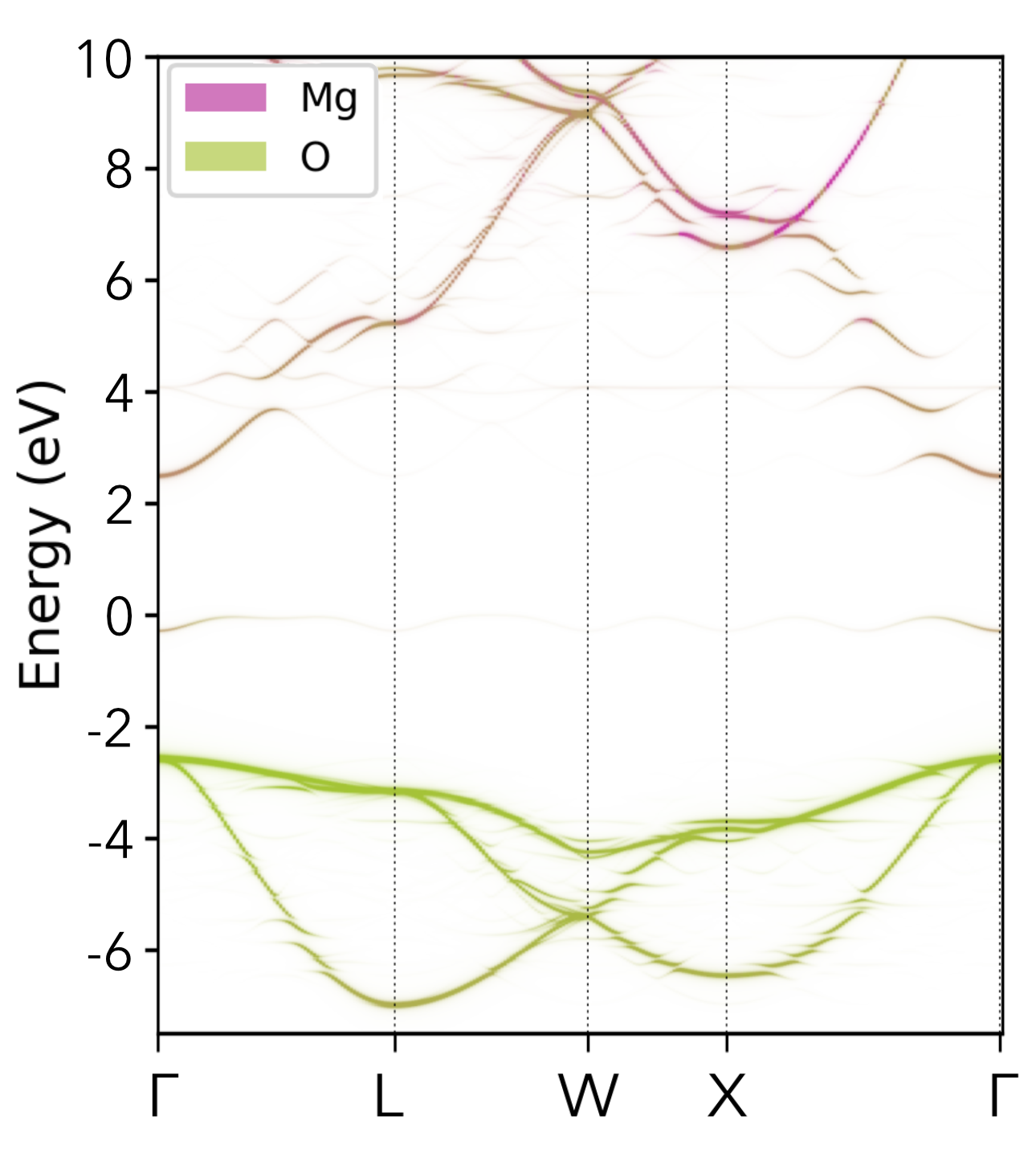
Oxygen Vacancy (Vₒ⁰) in MgO |
|
|---|---|
Tip
See the easyunfold YouTube tutorial for a quick overview of the theory of band structure unfolding, and a walkthrough of the calculation & analysis workflow with easyunfold.
Usage#
To generate an unfolded band structure, one typically needs to perform the following steps:
Create a primitive unit cell, and generate a band structure k-point path corresponding to this primitive cell.
Create a supercell (e.g. disordered, defective, surface slab etc.), and obtain its optimised structure.
Generate a series of k-points for the supercell to be calculated.
Perform a band structure calculation with the supercell, and save its wavefunction output to file.
Post-process the supercell wavefunction to obtain the unfolded band structure in the k-point path of the primitive unit cell.
These generation and analysis steps are automated in easyunfold, with only the primitive unit cell and
supercell structures required as inputs from the user.
Typically, the supercell comprises some form of symmetry-breaking relative to the primitive cell, such as defects, disorder (e.g. special quasi-random structures (SQS) for site disorder – other forms of disorder such as magnetic, dynamic/vibrational, polar, elastic etc. also possible), or a surface/interface slab. In all cases, the supercell symmetry is lowered compared to the pristine primitive cell. Hence, for a given k-point path in the primitive cell Brillouin Zone, additional k-points are required to be sampled for the supercell, and the extracted spectral weights need to be appropriately averaged to obtain the correct effective band structure (EBS). See the docs Theory page and/or JOSS paper for more details.
Citation#
If you use easyunfold in your work, please cite:
B. Zhu, S. R. Kavanagh & D. O. Scanlon, (2024). easyunfold: A Python package for unfolding electronic band structures. Journal of Open Source Software, 9(93), 5974, https://doi.org/10.21105/joss.05974
Studies using easyunfold#
We’ll add papers that use easyunfold to this list as they come out!
J. Tu et al. Giant switchable ferroelectric photovoltage in double-perovskite epitaxial films through chemical negative strain Science Advances 2025
L. F. Leon-Pinzon et al. Observation of Pseudogap in Cr1−xYxN magnetic alloy and its impact on the Seebeck coefficient by ab-initio calculations arXiv 2025
C. Zhang & J. Recatala-Gomez & Z. Aabdin & Y. Jiang et al. Direct Joule-Heated Non-Equilibrium Synthesis Enables High Performing Thermoelectrics arXiv 2025
W. Feng et al. Unraveling the role of oxygen vacancies in the electronic and optical properties of κ-Ga2O3 ResearchSquare 2025
J. J. Plata et al. High-Entropy Skutterudites as Thermoelectrics: Synthesizability and Band Convergence via the Cocktail Effect ChemRxiv 2025
P. B. Colominas et al. Giant Thermally Induced Band-Gap Renormalization in Anharmonic Silver Chalcohalide Antiperovskites Journal of Materials Chemistry C 2025
L. Zhang et al. Impurity point defects in Mg doping Al0.5Ga0.5N: A first principles study Computational Materials Science 2025
H. Maleki-Ghaleh et al. Visible Light-Sensitive Sustainable Quantum Dot Crystals of Co/Mg Doped Natural Hydroxyapatite Possessing Antimicrobial Activity and Biocompatibility Small 2024
L. Richarz et al. Ferroelectric domain walls for environmental sensors arXiv 2024
K. Eggestad, B. A. D. Williamson, D. Meier and S. M. Selbach Mobile Intrinsic Point Defects for Conductive Neutral Domain Walls in LiNbO3 Journal of Materials Chemistry C 2024
L. Zhang et al. Study of native point defects in Al0.5Ga0.5N by first principles calculations Computational Materials Science 2024
Dargahi et al. Synthesis and characterization of zinc-doped hematite nanoparticles for photocatalytic applications and their electronic structure studies by density functional theory Optical Materials 2024
S. M. Liga & S. R. Kavanagh, A. Walsh, D. O. Scanlon and G. Konstantatos Mixed-Cation Vacancy-Ordered Perovskites (Cs2Ti1–xSnxX6; X = I or Br): Low-Temperature Miscibility, Additivity, and Tunable Stability Journal of Physical Chemistry C 2023
Y. T. Huang & S. R. Kavanagh et al. Strong absorption and ultrafast localisation in NaBiS2 nanocrystals with slow charge-carrier recombination Nature Communications 2022
A. T. J. Nicolson et al. Interplay of Static and Dynamic Disorder in the Mixed-Metal Chalcohalide Sn2SbS2I3 Journal of the Americal Chemical Society 2023
Y. Wang & S. R. Kavanagh et al. Cation disorder engineering yields AgBiS2 nanocrystals with enhanced optical absorption for efficient ultrathin solar cells Nature Photonics 2022 (early version)
DFT code support#
At the moment, easyunfold supports VASP and CASTEP, but most of the routines are abstracted from the code specific details. In principle, support for other plane wave DFT code can be added by:
Implementing a subclass of
WaveFunctionthat handles reading the wave function output.Implementing functions for reading/writing k-points.
Adding branches for dispatching based on the
dft_codeattribute of theUnfoldKSetobject in various places within the code.
The Atomic Simulation Environment (ASE) is used by easyunfold for
reading in structures, so structure file IO is natively supported for essentially all public DFT codes.
In fact, ASE can already run band structure calculations using many plane-wave DFT codes. However, reading the plane wave coefficients from calculation outputs is not widely supported yet, which are needed here for band unfolding. Nevertheless, using ASE’s existing IO framework to widen the code support can be a fruitful direction for further development.
Code Compatibility Notes#
Atom-projected band structures are currently only supported for
VASPcalculation outputs.Gamma-only and non-collinear spin calculations are not supported for
CASTEP.
Contributors#
And those who helped in the development:
Bugs reports and feature requests#
Bug reports and feature requests are well come. If you found any bug or missing features please report it on the Issue Tracker.
Seeking support#
If you need support about using the software, please open a ticket with the help wanted label on the Issue Tracker.
Contributing#
Code contributions#
We welcome your help in improving and extending the package with your own contributions. This is managed through GitHub pull requests; for external contributions, we prefer the fork and pull workflow while core developers use branches in the main repository:
First open an Issue to discuss the proposed contribution. This discussion might include how the changes fit
easyunfold’s scope and a general technical approach.Make your own project fork and implement the changes there. Please keep your code style compliant and use the
pre-commithooks.Open a pull request to merge the changes into the main project. A more detailed discussion can take place there before the changes are accepted.
Development#
To develope the package, please clone it on GitHub and install it with the doc and test extras so all dependencies needed for development are installed:
git clone https://github.com/SMTG-Bham/easyunfold
pip install -e "./easyunfold[doc,test]"
To run the tests, simply run the pytest command while in the top-level code repository.
To build the documentation, run the make html command while in the docs directory.
If you want to add new features, please also include a test case to check that they work as expected. For more information please consult pytest’s documentation.
Please note that we use pre-commit hooks for code formatting and linting.
Please install them using pre-commit install so these hooks can run before committing code updates.