Si supercell using CASTEP#
Below is a step-by-step guide for unfolding a 2x1x1 pristine supercell sampled with a simple \(\Gamma - X\) path using CASTEP.
Generate the project file (easyunfold.json) and k-points for supercell calculation#
Note
The files needed for this example are provided in the examples/Si-castep folder. This guide assumes the current working directory is located at the root of that folder.
First, generate the supercell k-points:
easyunfold generate Si_prim.cell Si_211_band/Si_211_band.cell band.cell --code castep
Tip
Here, band.cell contains the band structure path and labels of the high symmetry k-points for the
primitive cell. The sumo package can be used to generate it with ease.
The easyunfold generate command above also generates an easyunfold.json file in the current directory,
containing information about the unfolding. The name of this file can be modified with the --out-file
command-line argument.
Information stored in this file can be inspected with easyunfold unfold status command:
$ easyunfold unfold status
Loaded data from easyunfold.json
Supercell cell information:
Space group number: 227
International symbol: Fd-3m
Point group: m-3m
Primitive cell information:
Space group number: 227
International symbol: Fd-3m
Point group: m-3m
No. of k points in the primitive cell : 21
No. of supercell k points : 51
No. of primitive cell symmetry operations : 48
No. of supercell symmetry operations : 24
Path in the primitive cell:
\Gamma : 1
X : 21
In this example, the path goes from \(\Gamma\) to \(X\).
Copy the generated cell file to the supercell calculation folder:
cp easyunfold_sc_kpoints.cell Si_211_unfold/
Perform the supercell band structure calculation#
Band structure calculations are performed in CASTEP by having the following keys in the <seed>.param
file:
task : spectral
spectral_task: bandstructure
write_orbitals : true
The last key is necessary for band unfolding, as it enables the wave function of the supercell band
structure calculation to be written to the disk in the <seed>.orbitals file.
Assuming CASTEP is installed, the following command will launch the calculation:
cd Si_211_unfold
mpirun -np 4 castep.mpi easyunfold_sc_kpoints
CASTEP will first perform a standard self-consistent field loop to find the ground state electron
density (and hence the potential). Following this, (in the same run) CASTEP switches to spectral
calculation and solves for the eigenvalues at each specified k-point with the electronic potential
(density) kept fixed.
Perform band unfolding#
Unfold the supercell wave function (.orbitals file) and calculate the spectral weights:
cd ../
easyunfold unfold calculate Si_211_unfold/easyunfold_sc_kpoints.orbitals
Plot the unfolded band structure:
easyunfold unfold plot
Output:
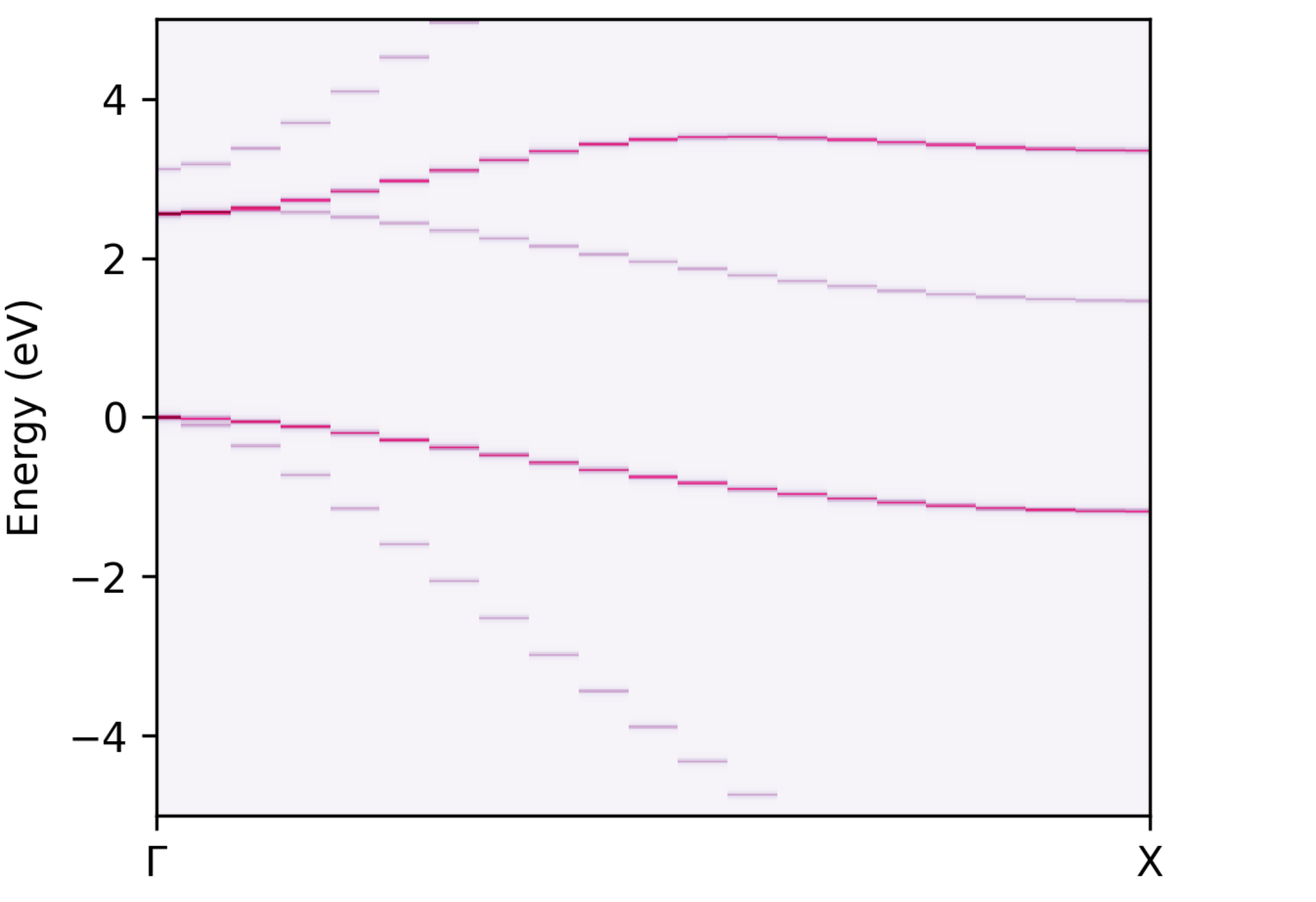
Spectral function of the unfolded bands.#
Tip
See the NaBiS2 example for tips on customising and prettifying the unfolded band structure plot.
Band structures of the primitive cell and supercell#
Below is the band structure of the primitive cell, along the same k-point path:
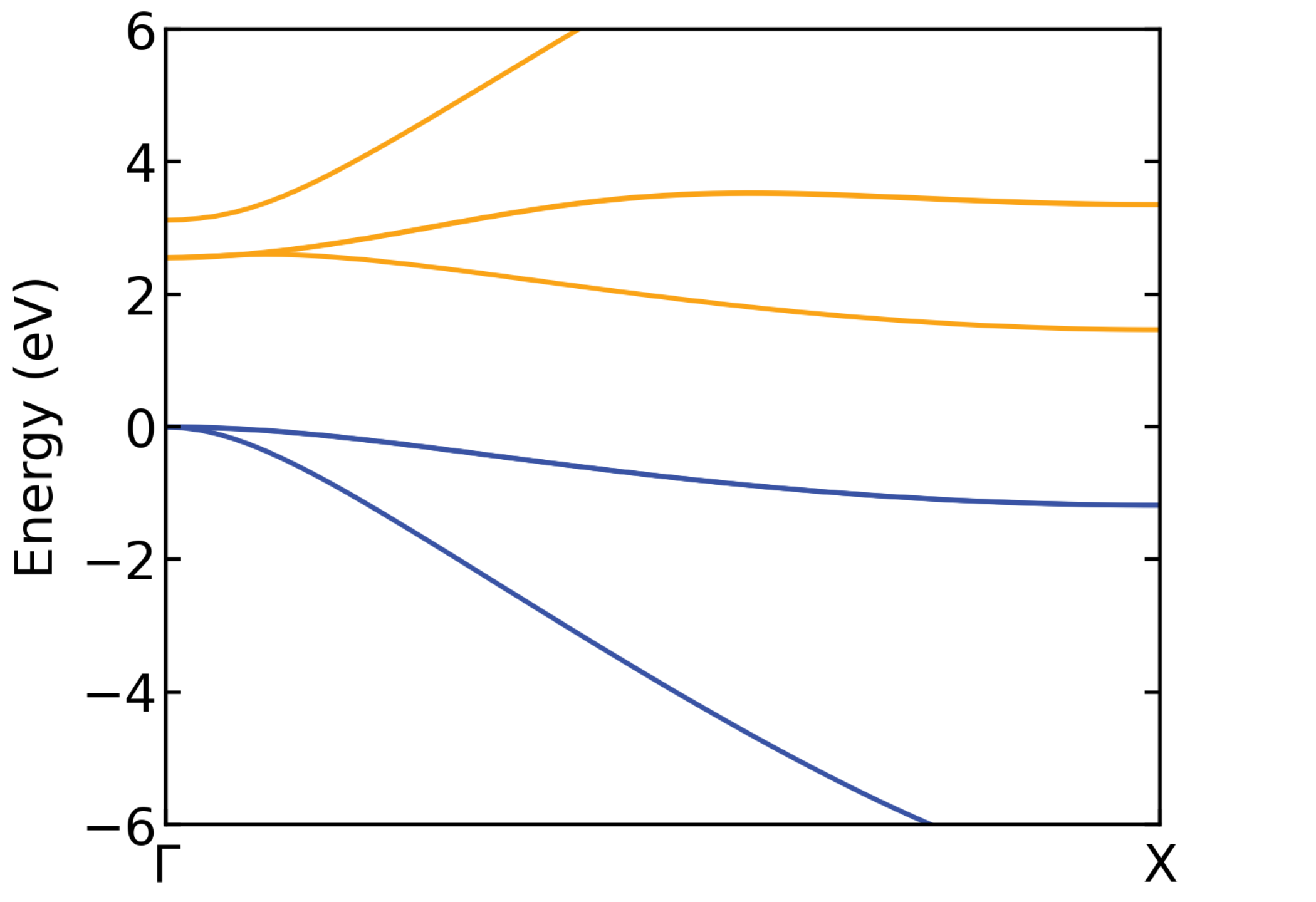
Primitive cell band structure of Si.#
which is well-matched by the unfolded band structure. This is not surprising, since a pristine supercell with no symmetry-breaking is used here.
Below is the band structure of the supercell that is obtained when blindly following the same k-point path (i.e. using the same k-point set) as the primitive cell:
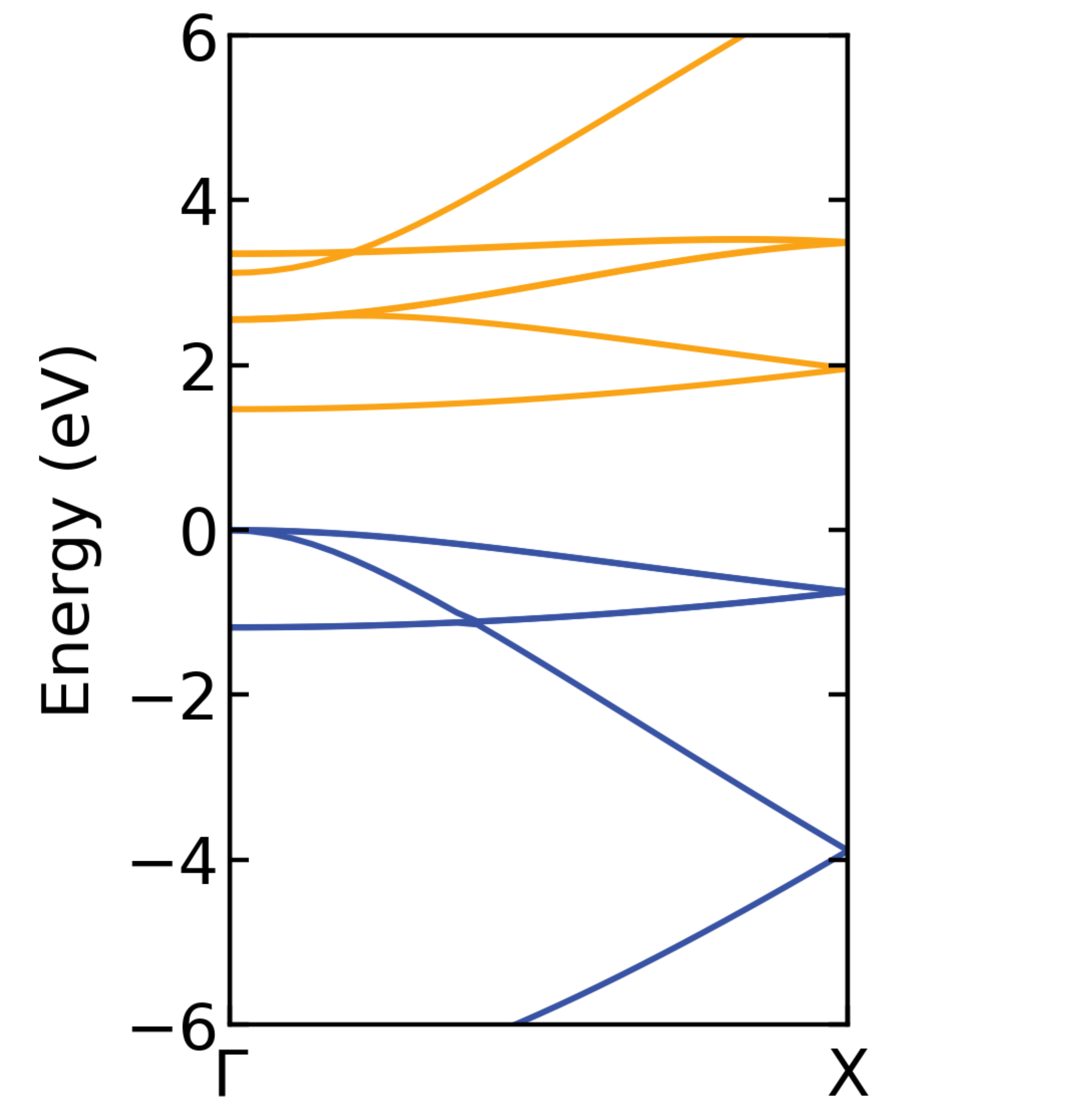
Supercell band structure of Si in a 2x1x1 supercell#
which, as expected, is the same as the primitive cell if it is folded back along the midpoint between \(\Gamma\) and \(X\), corresponding to the 2x expansion of the primitive cell along the \(x\) direction in generating the 2x1x1 supercell.
Tip
There are many customisation options available for the plotting functions in easyunfold. See easyunfold plot -h or
easyunfold unfold plot-projections -h for more details!