Heatmaps¶
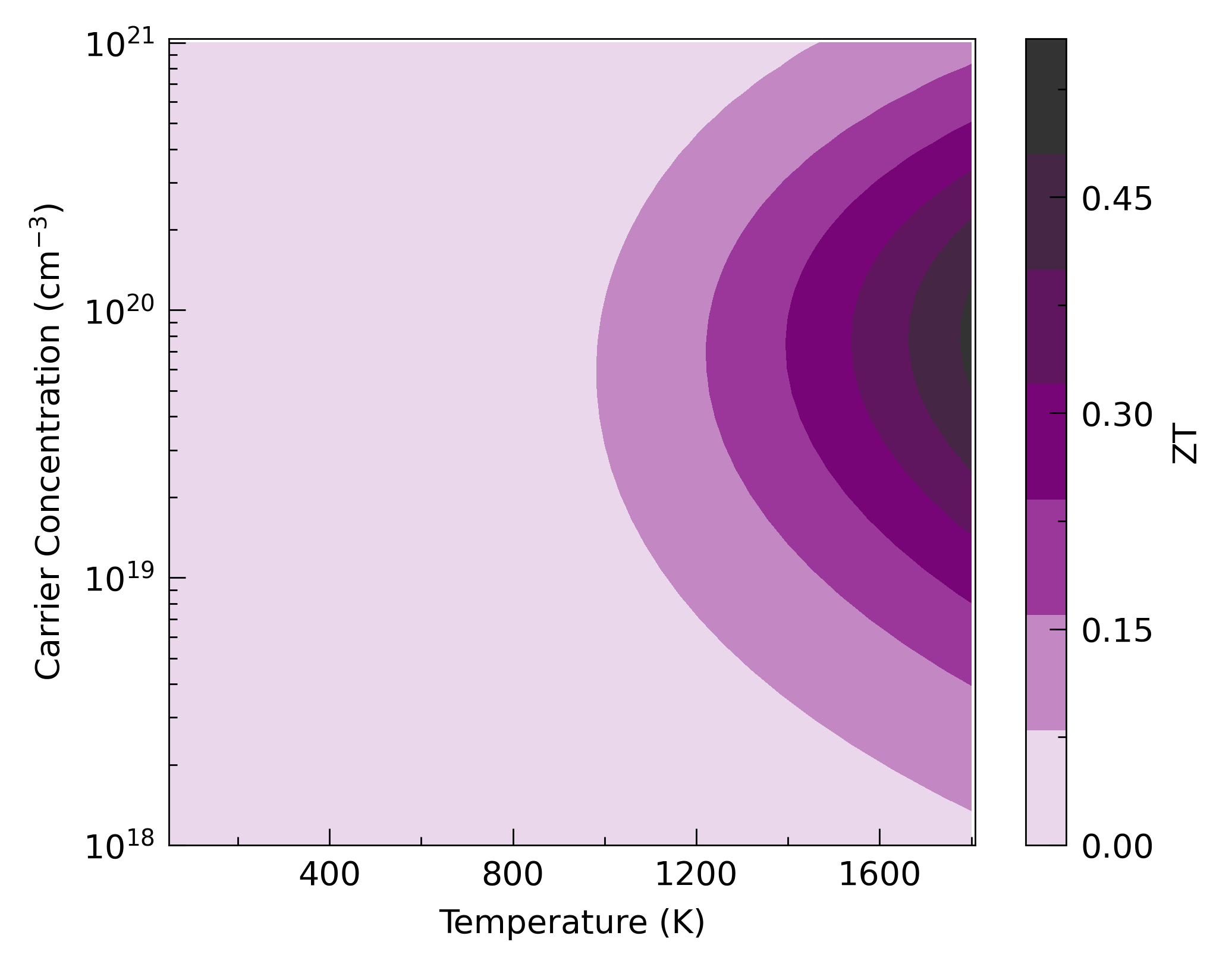
This shows the classic ZT against carrier concentration and temperature heatmap. This can be plotted at the command line with:
tp plot ztmap ../data/zno/boltztrap.hdf5 -k ../data/zno/kappa-m404021.hdf5 -c '#800080' -d x --discrete
and in python with:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 | #!/usr/bin/env python3
from os import path
import tp
bfile = '../data/zno/boltztrap.hdf5'
kfile = '../data/zno/kappa-m404021.hdf5'
if not path.isfile(kfile) or (path.getsize(kfile) < 1024*1024*100):
raise Exception('File not found, please use get-data.sh in the folder above.')
direction = 'x'
colour = '#800080'
# Axes
fig, ax, add_legend = tp.axes.small.one_colourbar()
# Load
adata = tp.data.load.boltztrap(bfile)
kdata = tp.data.load.phono3py(kfile)
# Add
tp.plot.heatmap.add_ztmap(ax, adata, kdata, direction=direction, colour=colour,
discrete=True)
# Save
fig.savefig('ztmap.pdf')
fig.savefig('ztmap.png')
|
This uses the tp.plot.heatmap.add_ztmap function (line 22), which
calculates the ZT and sets the axes labels. All the other functionality
is wrapped within the tp.plot.heatmap.add_heatmap function, which
enhances pcolourmesh in ways such as automatic rescaling of axes to
represent all data, automatic extension of colourbars if applicable and
custom colourmaps, which are generated dynamically with the input of a
single colour, in hex, rgb (array) or named colour format. In this
case, the tp.plot.colour.uniform colourmap generator has been used,
which calculates a uniform colourmap between white, a supplied
highlight colour and black, or three colours of your choosing. There
are several other heatmap functions in ThermoParser, including the
equivalent for power factor, add_pfmap, and those below…
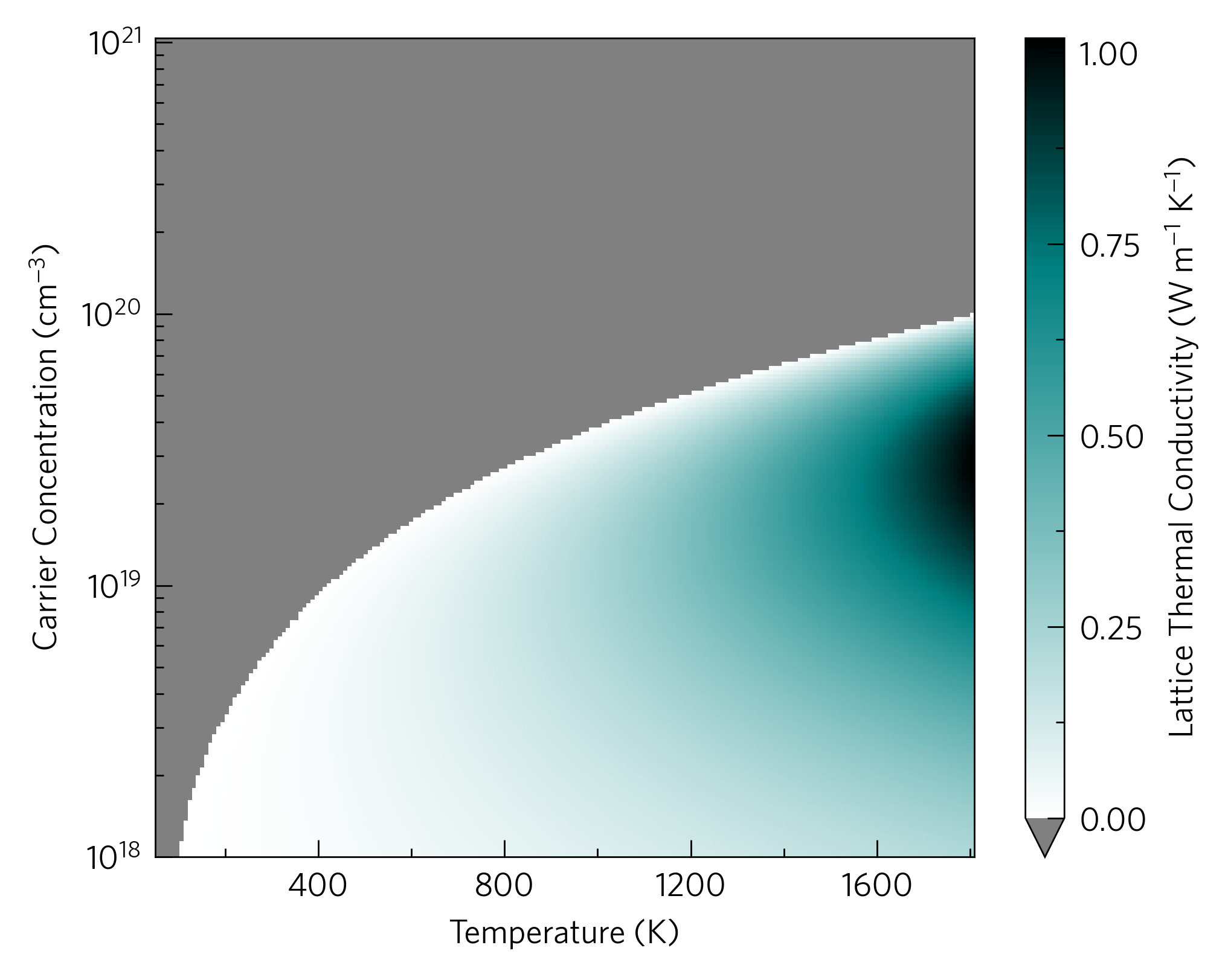
It is possible to plot a ztmap without lattice thermal conductivity
data, in which case it will be set to 1 W m<sup>-1</sup> K<sup>-1</sup>,
which will be recorded in data['meta']['kappa_source'] (other
numbers can be specified). The above is an alternative, which plots the
lattice thermal conductivity required to reach a specified ZT. We hope
this is a valuable tool to help decide wether to run expensive lattice
thermal conductivity calculations on a material. The CLI is:
tp plot kappa-target ../data/zno/boltztrap.hdf5 -c '#008080' -d x
and python:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 | #!/usr/bin/env python3
import tp
f = '../data/zno/boltztrap.hdf5'
target = 2
direction = 'x'
colour = tp.plot.colour.uniform('#008080')
# Axes
fig, ax, _ = tp.axes.small.one_colourbar()
# Load
data = tp.data.load.boltztrap(f)
# Add
tp.plot.heatmap.add_kappa_target(ax, data, zt=target, colour=colour,
direction=direction)
# Save
fig.savefig('kappa-target.pdf')
fig.savefig('kappa-target.png')
|
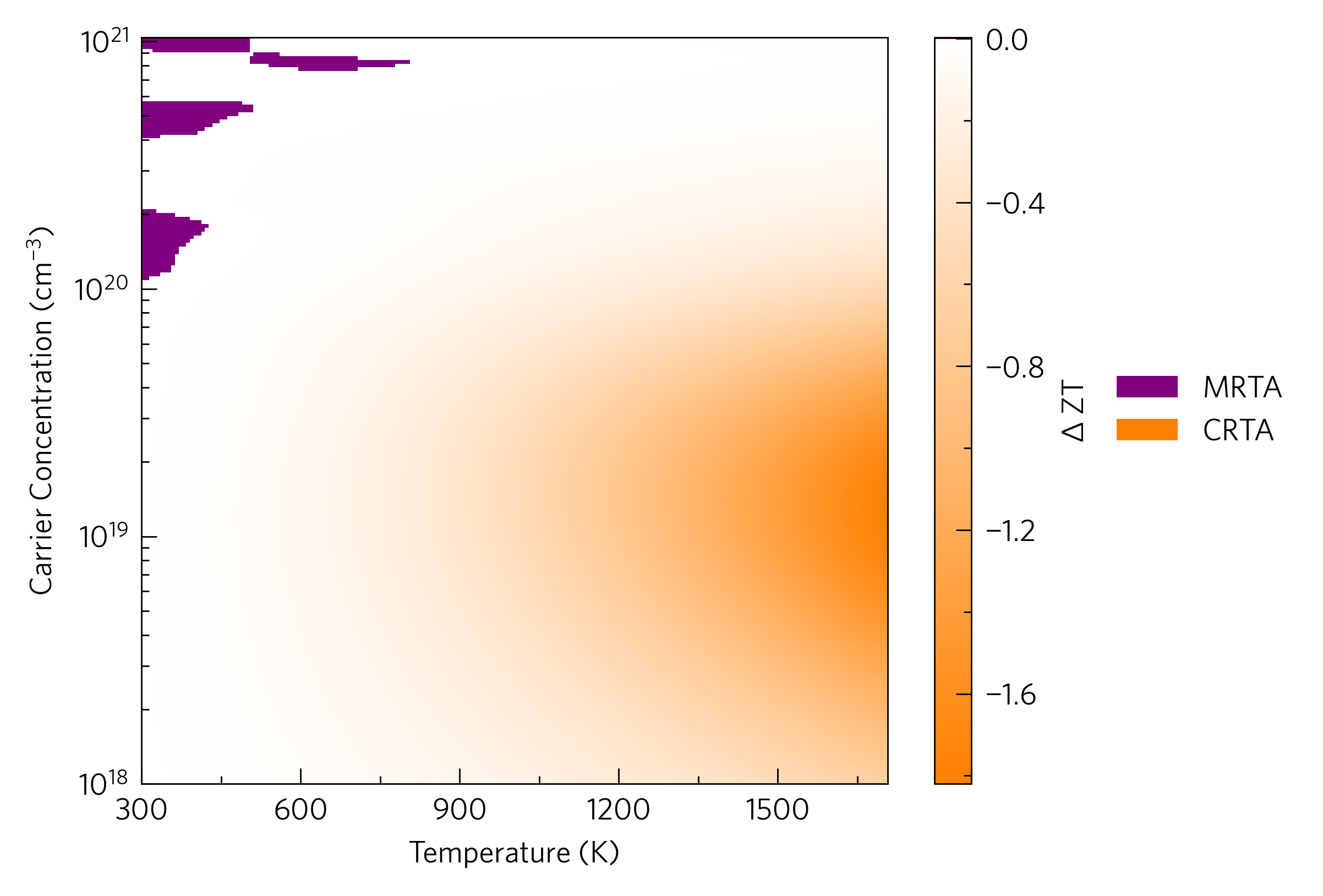
The final heatmap is the ztdiff, which compares the ZTs of two materials at a range of carrier concentrations and temperatures. This can be done with any two materials, from completely different to the same material under different conditions (e.g. different degrees of nanostructuring), although this method is most appropriate for closely related materials with similar dopabilities. Here we compare the momentum relaxation time approximation via AMSET with the constant relaxation approximation via BoltzTraP. At the command line:
tp plot ztdiff ../data/basno3/transport_75x75x75.json ../data/basno3/boltztrap.hdf5 -k ../data/basno3/kappa-m363636.hdf5 ../data/basno3/kappa-m363636.hdf5 -l{MRTA,CRTA}
and in python:
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 | #!/usr/bin/env python3
from os import path
import tp
efiles = ['../data/basno3/transport_75x75x75.json',
'../data/basno3/boltztrap.hdf5']
kfile = '../data/basno3/kappa-m363636.hdf5'
label = ['MRTA', 'CRTA']
if not path.isfile(kfile) or (path.getsize(kfile) < 1024*1024*100):
raise Exception('File not found, please use get-data.sh in the folder above.')
# Axes
fig, ax, add_legend = tp.axes.small.one_colourbar()
# Load
adata = [tp.data.load.amset(efiles[0]),
tp.data.load.boltztrap(efiles[1])]
kdata = tp.data.load.phono3py(kfile)
# Add
_, h, l = tp.plot.heatmap.add_ztdiff(ax, *adata, kdata, kdata, label1=label[0],
label2=label[1])
add_legend(handles=h, labels=l)
# Save
fig.savefig('ztdiff.pdf')
fig.savefig('ztdiff.png')
|
Note how list comprehensions and expansions (lines 19-20 and 23
respectively) can make the script tidier. On top of the colourbar (here
thrown away with _), this function returns the handles and labels
which must be put into the legend manually if you want it. If you are
using an axes template with multiple sets of axes, the custom
argument must be used in add_legend to enable manual manipulation
of the handles and labels. add_ztdiff and its power factor
equivalent, add_pfdiff, also make sure the colour scale is centred
on zero (i.e. where both are equal).
Some users may find adding contours with the contours argument or making
the colour scale discrete with the discrete argument more legible or
informative.