________
///// \\\\
\________/________________________________________________________________
|_____ : ___ \
| | | :| \ /
| | |___ ___ |___ |_____ ___ :|___/ ___ |___ ___ ___ |___ \
| | | | / \ | \ | | | / \:| / | | \ / / \ | \/
| | | | |___/ | | | | | |:| | | | \___ |___/ | \_____
| | | | \___ | | | | \___/:| \___|_ | \ \___ | :3.3.0\
|____________________________________:_______________________/____________:_____/
ThermoParser is a toolkit used to simplify the analysis of data produced by specialist materials science codes, centred around thermoelectrics, but also useful for anything pertaining to electronic and/ or phononic transport. ThermoParser is a Python library which contains functions for data retrieval, manipulation and plotting, which can be easily used with a little Python knowlege to generate a wide array of high-quality plots in only a few lines of code. ThermoParser also contains a suite of command-line tools which can retrieve specific data, save derived properties and plot graphs in a single command.
Click on the image to go to the gallery!
Installation¶
ThermoParser can easily be installed with git and pip:
git clone https://github.com/smtg-bham/ThermoParser.git
cd ThermoParser
pip install .
After installing, you may want to copy ThermoParser/tprc.yaml to
~/.config/tprc.yaml, if you want to set your own default axis
labels, unit conversions, default style sheets (two are provided),
other aesthetic alterations and more!
Mac¶
If installing on an m1 mac, you can’t currently pip install h5py, so a longer process is required:
Install brew
Install hdf5 with brew
python3 -m pip install cython numpybrew info hdf5to retrieve the path to your hdf5 installHDF5_DIR=YOUR_HDF5_PATH --no-build-isolation h5pygit clone https://github.com/smtg-bham/ThermoParser.gitcd ThermoParserpip install --user -e .
Using conda may circumvent this process.
Command Line Interface (CLI)¶
ThermoParser uses click, which has an easily navigable structure
from the command-line, detailed in the tutorials.
The most frequently useful commands are included in the CLI for maximum
ease, including the tp get functions, which verbosely retrieve data
from files which are normally tiresome and error-prone to navigate; and
most of the simplest plot-types available through the Python interface.
Python Interface¶
ThermoParser is designed to have four main stages:
- Axes:
Pick an axis layout from
tp.axes.
- Load:
Use the functions is
tp.data.loadto load the relevant data.
- Add:
Use functions in
tp.plotmodules to add graphs to the axes.
- Save:
Use
plt.savefigor equivalent to produce the figure.
As ThermoParser is dependent on matplotlib, each stage can be
substituted with bespoke code, e.g. using matplotlib.pyplot.subplots
or matplotlib.axes.Axes.scatter. These can still be supplemented
with ThermoParser helper functions, such as default labels which the
user can set in tp.settings, colourmap generators in
tp.plot.colour or legend helpers such as tp.axes.legend.alphabetise.
The best way to get a feel for ThermoParser is to see it in action: Take a look at our examples and tutorials. Currently supported codes are:
Current plotting modes are split into four areas.
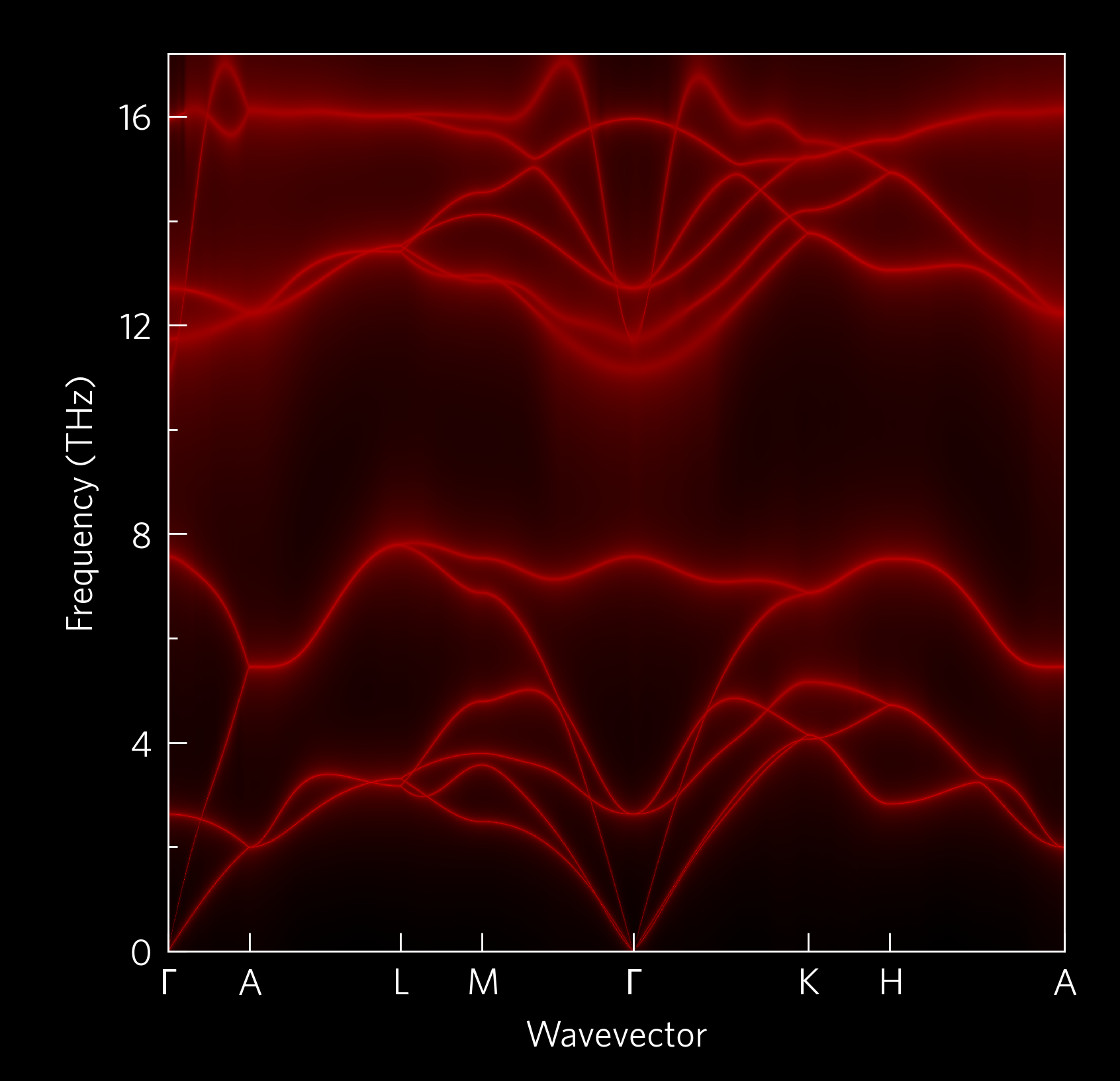
tp.plot.phononscontains plots along a high-symmetry path, including phonon dispersions and plots which project other quantities onto these paths in various ways.tp.plot.frequencyplots frequency on the x-axis, including density of states (DoS), cumulative kappa, “waterfall” and density plots. Each function has amainargument, which can be useful when plotting multiple quantities on the same set of axes; and aninvertargument, which swaps the x and y axes to let you plot DoS-style next to atp.plot.phononsplot.tp.plot.mfpcontains a cumulative kappa against mean free path plot.tp.plot.heatmapcontains a heatmap plotter, and wrappers which format appropriately for ZT against temperature and doping concentration; and one which plots the lattice thermal conductivity required to reach a target ZT, again against temperature and doping.
A set of example scripts is provided in the tp/examples folder and
in our online examples, and there is documentation.
Contributing¶
We welcome any contributions, whether they be a feature request or a new piece of code (or anything else). Adding options is inteded to be straightforward; the modularity of the code means that each step is mostly independent of the others.
Bugs and feature requests can be submitted to the issue tracker,
while contributions can be made using the fork and pull approach.
Contributions should include comprehensive docstrings, and where
appropriate examples, further documentation and tests are greatly
appreciated. Documentation uses the sphinx package, and can be built from
the docs directory with sphinx-build -b html src/ .. In order to build
the docs, download the extra dependencies with, e.g., pip install .[docs]
from the ThermoParser directory.
Testing¶
Tests use the unittest package, and can be run from the test directory
with python3 -m unittest. If you don’t already have unittest installed, it
can be directly with pip install unitest or, e.g., pip install .[tests]
from the ThermoParser directory.
Contributors¶
Many thanks to all those who contributed code or ideas to ThermoParser! Roughly chronologically, they are so far:
Kieran B. Spooner
Maud Einhorn
David O. Scanlon
Daniel W. Davies
Bonan Zhu
Sean R. Kavanagh
Warda Rahim
Katarina Brlec
Joe Willis
Dale Gaines II
Thanks also to the JOSS reviewers, Evan Walter Clarke Spotte-Smith, Enric Tomás Grau-Luque, and Francesco Nattino; and the editor Mojtaba Barzegari. An unintimidating and productive review process, which I would recommend if the opportunity arises!
License¶
ThermoParser is licensed under the GNU Affero General Public License v3 (AGPLv3).